Load packages and data
## Warning: package 'ggplot2' was built under R version 4.3.3
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
## Loaded dietaryindex
## Thank you for using dietaryindex!
## Tutorials: https://github.com/jamesjiadazhan/dietaryindex
## Dietary index calculations vary by the research question of the analysis. Currently, dietaryindex only supports simple scoring algorithm methods for all dietary indexes. Please use dietaryindex responsibly according to your research question. For more information, please refer to this resource: https://epi.grants.cancer.gov/hei/tools.html.
## Cite us: citation('dietaryindex') or
## Zhan JJ, Hodge RA, Dunlop AL, et al. Dietaryindex: a user-friendly and versatile R package for standardizing dietary pattern analysis in epidemiological and clinical studies. Am J Clin Nutr. Published online August 23, 2024. doi:10.1016/j.ajcnut.2024.08.021
# include COMPLEX SURVEY DESIGN #######################################
library(survey)
## Loading required package: grid
## Loading required package: Matrix
##
## Attaching package: 'Matrix'
## The following objects are masked from 'package:tidyr':
##
## expand, pack, unpack
## Loading required package: survival
## Warning: package 'survival' was built under R version 4.3.3
##
## Attaching package: 'survey'
## The following object is masked from 'package:graphics':
##
## dotchart
options(survey.lonely.psu = "adjust") #accounts for the lonely psu problem from subsetting survey data to small groups
# Load the data
data("DASH_trial")
data("PREDIMED_trial")
data("NHANES_20172018")
# set up working dictionary
setwd("/Users/james/Library/Mobile Documents/com~apple~CloudDocs/Desktop/Emory University - Ph.D./dietaryindex_package/NHANES_combined/dietaryindex_NHANES/data/NHANES_combined")
# Load the NHANES data from 2005 to 2018
## NHANES 2005-2006
load("NHANES_20052006.rda")
## NHANES 2007-2008
load("NHANES_20072008.rda")
## NHANES 2009-2010
load("NHANES_20092010.rda")
## NHANES 2011-2012
load("NHANES_20112012.rda")
## NHANES 2013-2014
load("NHANES_20132014.rda")
## NHANES 2015-2016
load("NHANES_20152016.rda")
Dietary index calculations
# DASHI from the DASH trial
DASHI_DASH = DASHI(
SERV_DATA = DASH_trial,
RESPONDENTID = DASH_trial$Diet_Type,
TOTALKCAL_DASHI = DASH_trial$Kcal,
TOTAL_FAT_DASHI = DASH_trial$Totalfat_Percent,
SAT_FAT_DASHI = DASH_trial$Satfat_Percent,
PROTEIN_DASHI = DASH_trial$Protein_Percent,
CHOLESTEROL_DASHI = DASH_trial$Cholesterol,
FIBER_DASHI = DASH_trial$Fiber,
POTASSIUM_DASHI = DASH_trial$Potassium,
MAGNESIUM_DASHI = DASH_trial$Magnesium,
CALCIUM_DASHI = DASH_trial$Calcium,
SODIUM_DASHI = DASH_trial$Sodium)
# MEDI for the PREDIMED trial
MEDI_PREDIMED = MEDI(
SERV_DATA = PREDIMED_trial,
RESPONDENTID = PREDIMED_trial$Diet_Type,
OLIVE_OIL_SERV_MEDI = PREDIMED_trial$Virgin_Oliveoil,
FRT_SERV_MEDI = PREDIMED_trial$Fruits,
VEG_SERV_MEDI = PREDIMED_trial$Vegetables,
LEGUMES_SERV_MEDI = PREDIMED_trial$Legumes,
NUTS_SERV_MEDI = PREDIMED_trial$Total_nuts,
FISH_SEAFOOD_SERV_MEDI = PREDIMED_trial$Fish_Seafood,
ALCOHOL_SERV_MEDI = PREDIMED_trial$Alcohol,
SSB_SERV_MEDI = PREDIMED_trial$Soda_Drinks,
SWEETS_SERV_MEDI = PREDIMED_trial$Sweets,
DISCRET_FAT_SERV_MEDI = PREDIMED_trial$Refined_Oliveoil,
REDPROC_MEAT_SERV_MEDI = PREDIMED_trial$Meat)
# set up survey design for NHANES data 2017-2018 day 1 and day 2
## filter out the missing values for the weight variable WTDR2D
## select only persons aged two and older
NHANES_20172018_design_d1d2 = NHANES_20172018$FPED %>%
filter(!is.na(WTDR2D)) %>%
filter(RIDAGEYR >= 2)
# filter only the individuals with reliable recall status for FPED, FPED_IND, NUTRIENT, NUTRIENT_IND, FPED2, FPED_IND2, NUTRIENT2, NUTRIENT_IND2
## DAY 1: DR1DRSTZ == 1
## DAY 2: DR2DRSTZ == 1
# also, filter out those kcal intake < 500 or > 5000
## DAY 1: DR1TKCAL >= 500 & DR1TKCAL <= 5000
## DAY 2: DR2TKCAL >= 500 & DR2TKCAL <= 5000
NHANES_20172018$FPED_IND = NHANES_20172018$FPED_IND %>%
filter(DR1DRSTZ == 1)
NHANES_20172018$NUTRIENT_IND = NHANES_20172018$NUTRIENT_IND %>%
filter(DR1DRSTZ == 1)
NHANES_20172018$FPED = NHANES_20172018$FPED %>%
filter(DR1DRSTZ == 1)
NHANES_20172018$NUTRIENT = NHANES_20172018$NUTRIENT %>%
filter(DR1DRSTZ == 1) %>%
filter(DR1TKCAL >= 500 & DR1TKCAL <= 5000)
NHANES_20172018$FPED_IND2 = NHANES_20172018$FPED_IND2 %>%
filter(DR2DRSTZ == 1)
NHANES_20172018$NUTRIENT_IND2 = NHANES_20172018$NUTRIENT_IND2 %>%
filter(DR2DRSTZ == 1)
NHANES_20172018$FPED2 = NHANES_20172018$FPED2 %>%
filter(DR2DRSTZ == 1)
NHANES_20172018$NUTRIENT2 = NHANES_20172018$NUTRIENT2 %>%
filter(DR2DRSTZ == 1) %>%
filter(DR2TKCAL >= 500 & DR2TKCAL <= 5000)
## NHANES 2017-2018
# DASHI for day 1 and day 2
DASHI_NHANES = DASHI_NHANES_FPED(NUTRIENT_PATH=NHANES_20172018$NUTRIENT, NUTRIENT_PATH2=NHANES_20172018$NUTRIENT2)
# MEDI for day 1 and day 2
MEDI_NHANES = MEDI_NHANES_FPED(FPED_IND_PATH=NHANES_20172018$FPED_IND, NUTRIENT_IND_PATH=NHANES_20172018$NUTRIENT_IND, FPED_IND_PATH2=NHANES_20172018$FPED_IND2, NUTRIENT_IND_PATH2=NHANES_20172018$NUTRIENT_IND2)
## Since no SSB code is provided, the default SSB code from 17-18 FNDDS file is used.
## Since no SWEETS code is provided, the default SSB code from 17-18 FNDDS file is used
## Since no FAT_OIL code is provided, the default FAT_OIL code from 17-18 FNDDS file is used
## Since no OLIVE_OIL code is provided, the default OLIVE_OIL code from 17-18 FNDDS file is used
# DASH for day 1 and day 2
DASH_NHANES = DASH_NHANES_FPED(NHANES_20172018$FPED_IND, NHANES_20172018$NUTRIENT_IND, NHANES_20172018$FPED_IND2, NHANES_20172018$NUTRIENT_IND2)
## Since no SSB code is provided, the default SSB code from 17-18 FNDDS file is used.
## Since no skim milk code is provided, the default skim milk code from 17-18 FNDDS file is used.
## Since no low-fat cheese code is provided, the default low-fat cheese code from 17-18 FNDDS file is used.
## Reminder: this DASH index uses quintiles to rank participants' food/drink serving sizes and then calculate DASH component scores, which may generate results that are specific to your study population but not comparable to other populations.
# MED for day 1 and day 2
MED_NHANES = MED_NHANES_FPED(FPED_PATH=NHANES_20172018$FPED, NUTRIENT_PATH=NHANES_20172018$NUTRIENT, DEMO_PATH=NHANES_20172018$DEMO, FPED_PATH2=NHANES_20172018$FPED2, NUTRIENT_PATH2=NHANES_20172018$NUTRIENT2)
## Reminder: this MED index uses medians to rank participants' food/drink serving sizes and then calculate MED component scores, which may generate results that are specific to your study population but not comparable to other populations.
# AHEI for day 1 and day 2
AHEI_NHANES = AHEI_NHANES_FPED(NHANES_20172018$FPED_IND, NHANES_20172018$NUTRIENT_IND, NHANES_20172018$FPED_IND2, NHANES_20172018$NUTRIENT_IND2)
## Since no SSB code is provided, the default SSB code from 17-18 FNDDS file is used.
## Trans fat is not avaiable for NHANES, so it is not included in the AHEI score.
# DII for day 1 and day 2
DII_NHANES = DII_NHANES_FPED(FPED_PATH=NHANES_20172018$FPED, NUTRIENT_PATH=NHANES_20172018$NUTRIENT, DEMO_PATH=NHANES_20172018$DEMO, FPED_PATH2=NHANES_20172018$FPED2, NUTRIENT_PATH2=NHANES_20172018$NUTRIENT2)
## VITD is included in the calculation in the first day of NHANES data.
## VITD is included in the calculation in the second day of NHANES data.
## Reminder: This function does not use all the original DII variables. Eugenol, garlic, ginger, onion, trans fat, turmeric, Green/black tea, Flavan-3-ol, Flavones, Flavonols, Flavonones, Anthocyanidins, Isoflavones, Pepper, Thyme/oregano, Rosemary are not included because they are not available in NHANES.
## Day 1 and Day 2 data are used for the calculation.
# HEI2020 for day 1 and day 2
HEI2020_NHANES_1718 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20172018$FPED, NUTRIENT_PATH=NHANES_20172018$NUTRIENT, DEMO_PATH=NHANES_20172018$DEMO, FPED_PATH2=NHANES_20172018$FPED2, NUTRIENT_PATH2=NHANES_20172018$NUTRIENT2)
# Merge all the previous data frames into one data frame by SEQN
NHANES_20172018_dietaryindex_d1d2 = inner_join(NHANES_20172018_design_d1d2, DASHI_NHANES, by = "SEQN") %>%
inner_join(MEDI_NHANES, by = "SEQN") %>%
inner_join(DASH_NHANES, by = "SEQN") %>%
inner_join(MED_NHANES, by = "SEQN") %>%
inner_join(AHEI_NHANES, by = "SEQN") %>%
inner_join(DII_NHANES, by = "SEQN") %>%
inner_join(HEI2020_NHANES_1718, by = "SEQN")
# set up survey design for NHANES data 2017-2018 day 1 and day 2
NHANES_design_1718_d1d2 <- svydesign(
id = ~SDMVPSU,
strata = ~SDMVSTRA,
weight = ~WTDR2D,
data = NHANES_20172018_dietaryindex_d1d2, #set up survey design on the full dataset #can restrict at time of analysis
nest = TRUE)
# generate the svymean object for DASHI_ALL
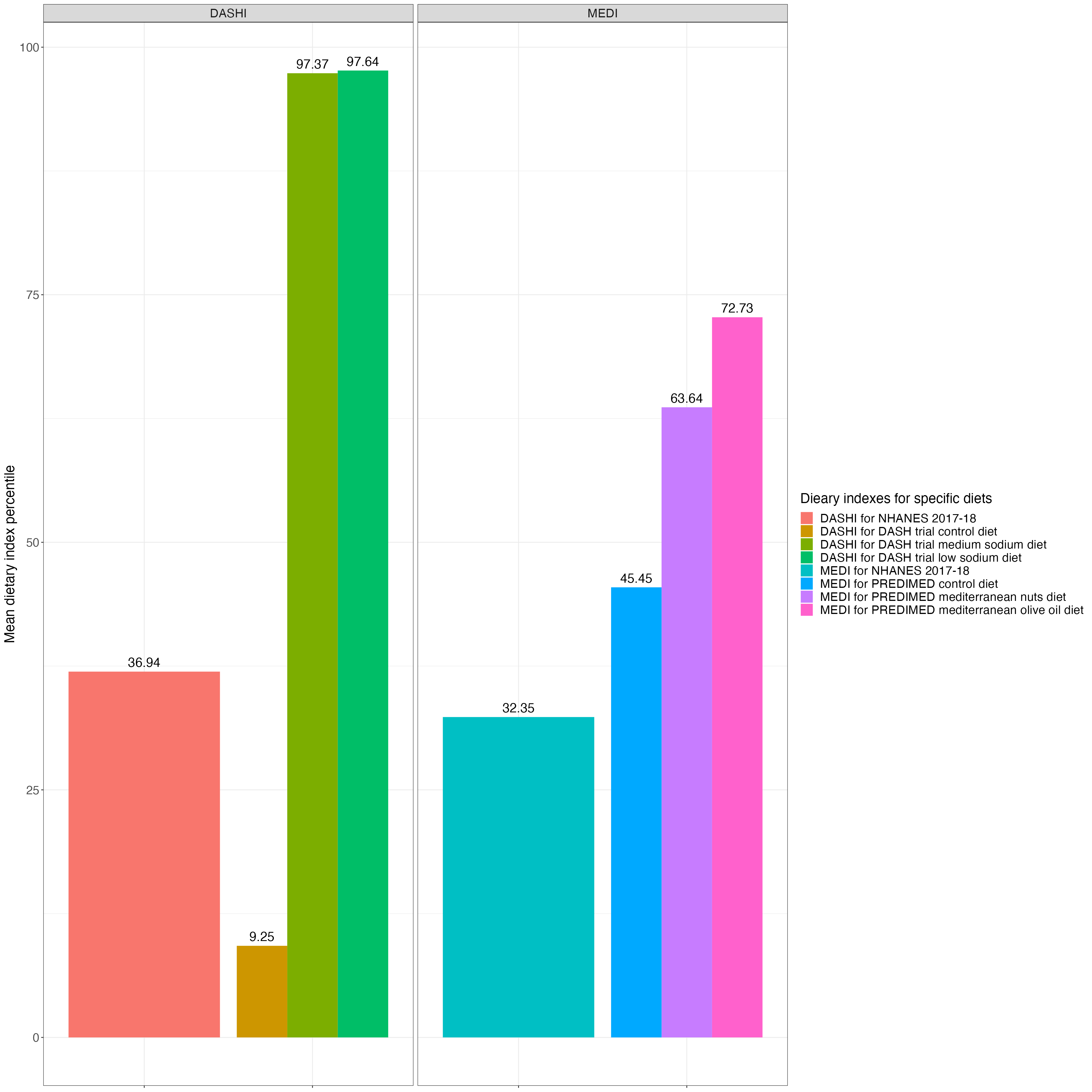
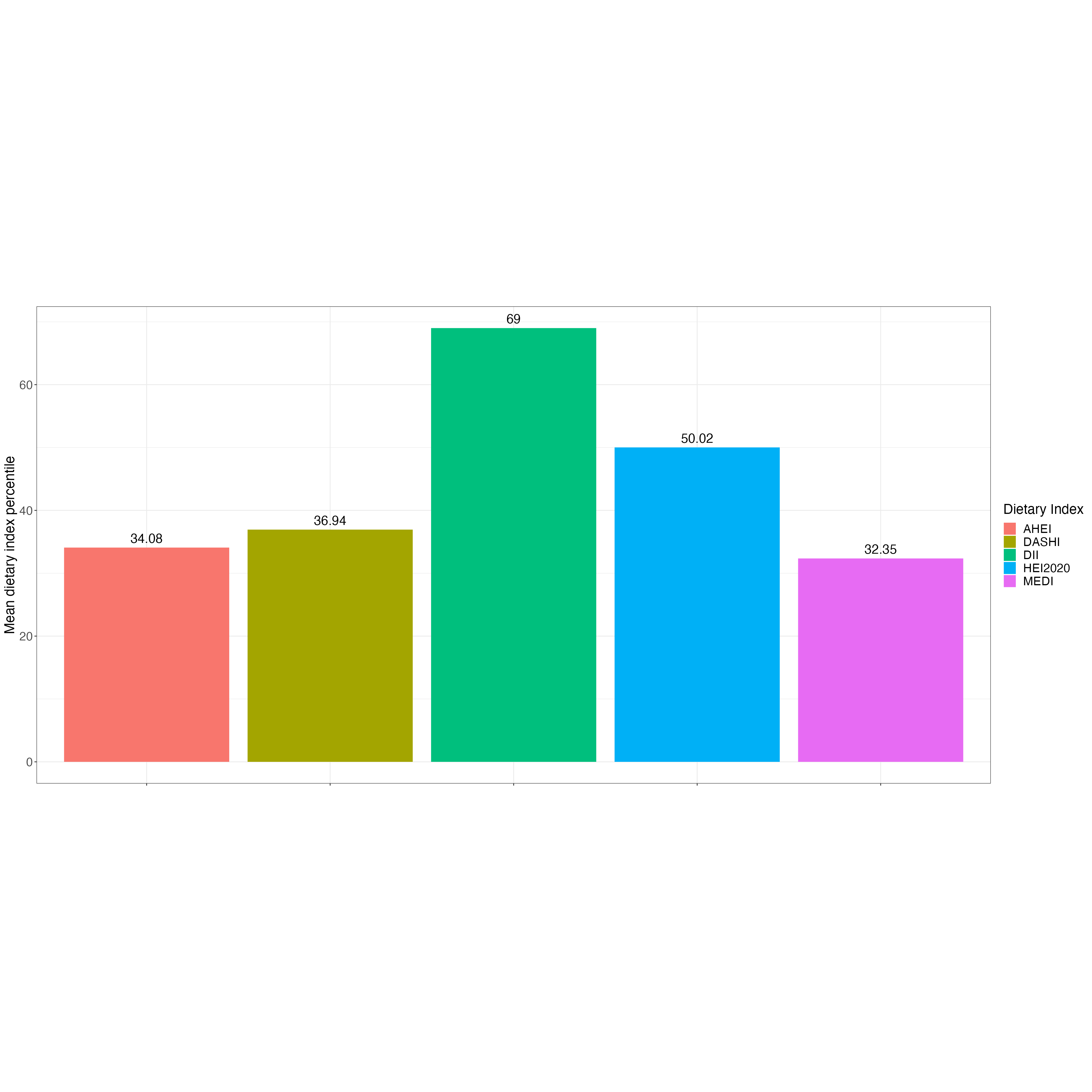
DASHI_1718_svymean = svymean(~DASHI_ALL, design = NHANES_design_1718_d1d2, na.rm = TRUE)
# extract the mean from the svymean object
DASHI_1718_svymean_mean = DASHI_1718_svymean[["DASHI_ALL"]]
# generate the svymean object for MEDI_ALL
MEDI_1718_svymean = svymean(~MEDI_ALL, design = NHANES_design_1718_d1d2, na.rm = TRUE)
# extract the mean from the svymean object
MEDI_1718_svymean_mean = MEDI_1718_svymean[["MEDI_ALL"]]
# generate the svymean object for DASH_ALL
DASH_1718_svymean = svymean(~DASH_ALL, design = NHANES_design_1718_d1d2, na.rm = TRUE)
# extract the mean from the svymean object
DASH_1718_svymean_mean = DASH_1718_svymean[["DASH_ALL"]]
# generate the svymean object for MED_ALL
MED_1718_svymean = svymean(~MED_ALL, design = NHANES_design_1718_d1d2, na.rm = TRUE)
# extract the mean from the svymean object
MED_1718_svymean_mean = MED_1718_svymean[["MED_ALL"]]
# generate the svymean object for AHEI_ALL
AHEI_1718_svymean = svymean(~AHEI_ALL, design = NHANES_design_1718_d1d2, na.rm = TRUE)
# extract the mean from the svymean object
AHEI_1718_svymean_mean = AHEI_1718_svymean[["AHEI_ALL"]]
# generate the svymean object for DII_ALL
DII_1718_svymean = svymean(~DII_ALL, design = NHANES_design_1718_d1d2, na.rm = TRUE)
# extract the mean from the svymean object
DII_1718_svymean_mean = DII_1718_svymean[["DII_ALL"]]
# generate the svymean object for HEI2020_ALL
HEI2020_1718_svymean = svymean(~HEI2020_ALL, design = NHANES_design_1718_d1d2, na.rm = TRUE)
# extract the mean from the svymean object
HEI2020_1718_svymean_mean = HEI2020_1718_svymean[["HEI2020_ALL"]]
# HEI2020 for day 1
## 2017-2018
HEI2020_NHANES_1718_d1 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20172018$FPED, NUTRIENT_PATH=NHANES_20172018$NUTRIENT, DEMO_PATH=NHANES_20172018$DEMO)
## add year column
HEI2020_NHANES_1718_d1 = HEI2020_NHANES_1718_d1 %>%
mutate(year = "2017-2018")
# select only necessary columns from FPED data
NHANES_20172018_FPED = NHANES_20172018$FPED %>%
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1)
## inner join with the NHANES 2017-2018 day 1 FPED data
HEI2020_NHANES_1718_d1 = inner_join(NHANES_20172018_FPED, HEI2020_NHANES_1718_d1, by = "SEQN")
# perform data filtering for 2005-2016 NHANES data
# filter only the individuals with reliable recall status for FPED, FPED_IND, NUTRIENT, NUTRIENT_IND, FPED2, FPED_IND2, NUTRIENT2, NUTRIENT_IND2
## DAY 1: DR1DRSTZ == 1
# also, filter out those kcal intake < 500 or > 5000
## DAY 1: DR1TKCAL >= 500 & DR1TKCAL <= 5000
## 2015-2016
NHANES_20152016$FPED = NHANES_20152016$FPED %>%
filter(DR1DRSTZ == 1)
NHANES_20152016$NUTRIENT = NHANES_20152016$NUTRIENT %>%
filter(DR1DRSTZ == 1) %>%
filter(DR1TKCAL >= 500 & DR1TKCAL <= 5000)
## 2013-2014
NHANES_20132014$FPED = NHANES_20132014$FPED %>%
filter(DR1DRSTZ == 1)
NHANES_20132014$NUTRIENT = NHANES_20132014$NUTRIENT %>%
filter(DR1DRSTZ == 1) %>%
filter(DR1TKCAL >= 500 & DR1TKCAL <= 5000)
## 2011-2012
NHANES_20112012$FPED = NHANES_20112012$FPED %>%
filter(DR1DRSTZ == 1)
NHANES_20112012$NUTRIENT = NHANES_20112012$NUTRIENT %>%
filter(DR1DRSTZ == 1) %>%
filter(DR1TKCAL >= 500 & DR1TKCAL <= 5000)
## 2009-2010
NHANES_20092010$FPED = NHANES_20092010$FPED %>%
filter(DR1DRSTZ == 1)
NHANES_20092010$NUTRIENT = NHANES_20092010$NUTRIENT %>%
filter(DR1DRSTZ == 1) %>%
filter(DR1TKCAL >= 500 & DR1TKCAL <= 5000)
## 2007-2008
NHANES_20072008$FPED = NHANES_20072008$FPED %>%
filter(DR1DRSTZ == 1)
NHANES_20072008$NUTRIENT = NHANES_20072008$NUTRIENT %>%
filter(DR1DRSTZ == 1) %>%
filter(DR1TKCAL >= 500 & DR1TKCAL <= 5000)
## 2005-2006
NHANES_20052006$FPED = NHANES_20052006$FPED %>%
filter(DR1DRSTZ == 1)
NHANES_20052006$NUTRIENT = NHANES_20052006$NUTRIENT %>%
filter(DR1DRSTZ == 1) %>%
filter(DR1TKCAL >= 500 & DR1TKCAL <= 5000)
## 2015-2016
HEI2020_NHANES_1516_d1 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20152016$FPED, NUTRIENT_PATH=NHANES_20152016$NUTRIENT, DEMO_PATH=NHANES_20152016$DEMO)
## add year column
HEI2020_NHANES_1516_d1 = HEI2020_NHANES_1516_d1 %>%
mutate(year = "2015-2016")
# select only necessary columns from FPED data
NHANES_20152016_FPED = NHANES_20152016$FPED %>%
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1)
## inner join with the NHANES 2015-2016 day 1 FPED data
HEI2020_NHANES_1516_d1 = inner_join(NHANES_20152016_FPED, HEI2020_NHANES_1516_d1, by = "SEQN")
## 2013-2014
HEI2020_NHANES_1314_d1 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20132014$FPED, NUTRIENT_PATH=NHANES_20132014$NUTRIENT, DEMO_PATH=NHANES_20132014$DEMO)
## add year column
HEI2020_NHANES_1314_d1 = HEI2020_NHANES_1314_d1 %>%
mutate(year = "2013-2014")
# select only necessary columns from FPED data
NHANES_20132014_FPED = NHANES_20132014$FPED %>%
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1)
## inner join with the NHANES 2013-2014 day 1 FPED data
HEI2020_NHANES_1314_d1 = inner_join(NHANES_20132014_FPED, HEI2020_NHANES_1314_d1, by = "SEQN")
## 2011-2012
HEI2020_NHANES_1112_d1 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20112012$FPED, NUTRIENT_PATH=NHANES_20112012$NUTRIENT, DEMO_PATH=NHANES_20112012$DEMO)
## add year column
HEI2020_NHANES_1112_d1 = HEI2020_NHANES_1112_d1 %>%
mutate(year = "2011-2012")
# select only necessary columns from FPED data
NHANES_20112012_FPED = NHANES_20112012$FPED %>%
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1)
## inner join with the NHANES 2011-2012 day 1 FPED data
HEI2020_NHANES_1112_d1 = inner_join(NHANES_20112012_FPED, HEI2020_NHANES_1112_d1, by = "SEQN")
## 2009-2010
HEI2020_NHANES_0910_d1 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20092010$FPED, NUTRIENT_PATH=NHANES_20092010$NUTRIENT, DEMO_PATH=NHANES_20092010$DEMO)
## add year column
HEI2020_NHANES_0910_d1 = HEI2020_NHANES_0910_d1 %>%
mutate(year = "2009-2010")
# select only necessary columns from FPED data
NHANES_20092010_FPED = NHANES_20092010$FPED %>%
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1)
## inner join with the NHANES 2009-2010 day 1 FPED data
HEI2020_NHANES_0910_d1 = inner_join(NHANES_20092010_FPED, HEI2020_NHANES_0910_d1, by = "SEQN")
## 2007-2008
HEI2020_NHANES_0708_d1 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20072008$FPED, NUTRIENT_PATH=NHANES_20072008$NUTRIENT, DEMO_PATH=NHANES_20072008$DEMO)
## add year column
HEI2020_NHANES_0708_d1 = HEI2020_NHANES_0708_d1 %>%
mutate(year = "2007-2008")
# select only necessary columns from FPED data
NHANES_20072008_FPED = NHANES_20072008$FPED %>%
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1)
## inner join with the NHANES 2007-2008 day 1 FPED data
HEI2020_NHANES_0708_d1 = inner_join(NHANES_20072008_FPED, HEI2020_NHANES_0708_d1, by = "SEQN")
## 2005-2006
HEI2020_NHANES_0506_d1 = HEI2020_NHANES_FPED(FPED_PATH=NHANES_20052006$FPED, NUTRIENT_PATH=NHANES_20052006$NUTRIENT, DEMO_PATH=NHANES_20052006$DEMO)
## add year column
HEI2020_NHANES_0506_d1 = HEI2020_NHANES_0506_d1 %>%
mutate(year = "2005-2006")
# select only necessary columns from FPED data
NHANES_20052006_FPED = NHANES_20052006$FPED %>%
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1)
## inner join with the NHANES 2005-2006 day 1 FPED data
HEI2020_NHANES_0506_d1 = inner_join(NHANES_20052006_FPED, HEI2020_NHANES_0506_d1, by = "SEQN")
# combine all the HEI2020 day 1 data by rows
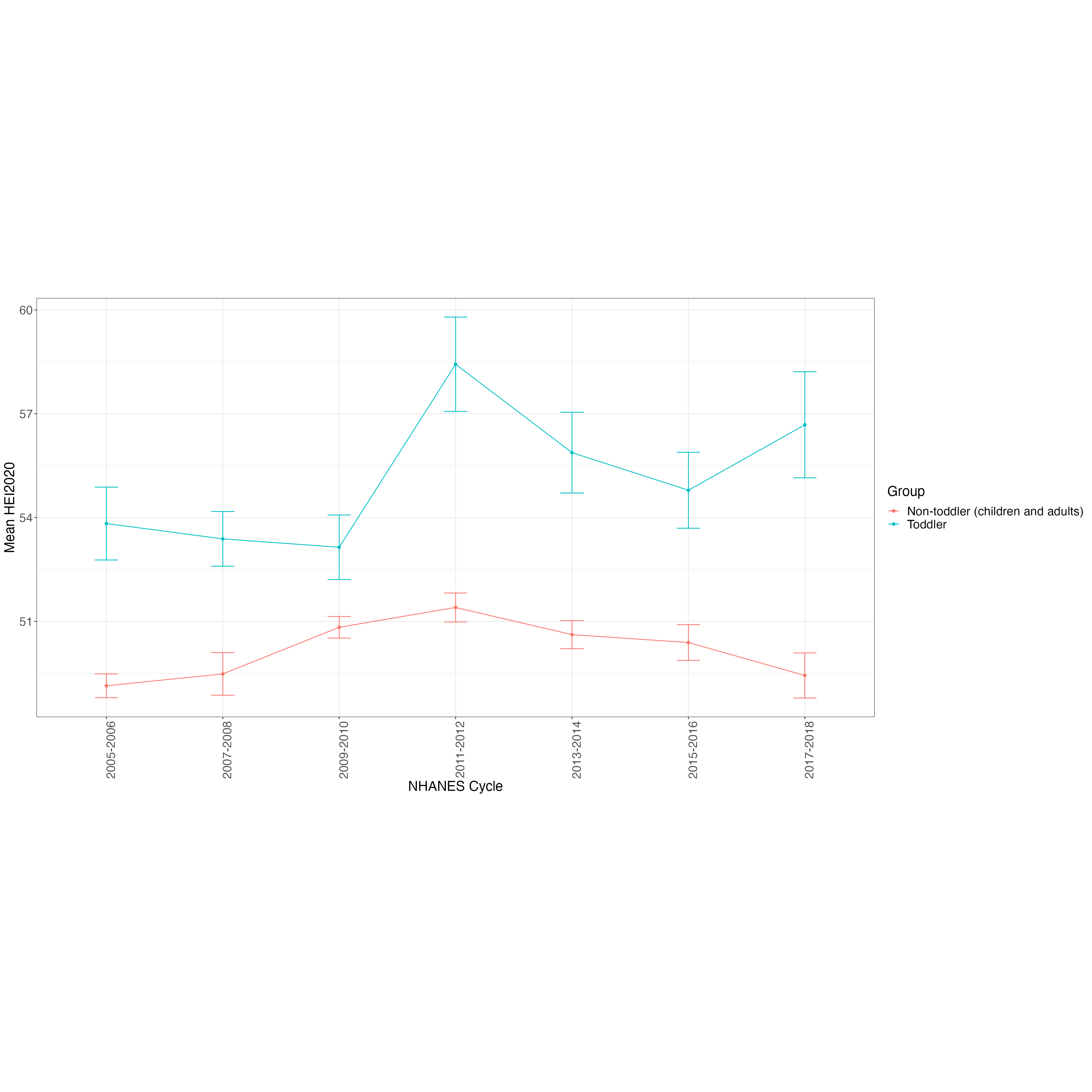
HEI2020_NHANES_d1 = rbind(HEI2020_NHANES_0506_d1, HEI2020_NHANES_0708_d1, HEI2020_NHANES_0910_d1, HEI2020_NHANES_1112_d1, HEI2020_NHANES_1314_d1, HEI2020_NHANES_1516_d1, HEI2020_NHANES_1718_d1)
# add toddler column
HEI2020_NHANES_d1_2 = HEI2020_NHANES_d1 %>%
mutate(toddler = ifelse(RIDAGEYR < 2, "toddler", "nontoddler")) %>%
## rearrange the column order
select(SEQN, SDMVPSU, SDMVSTRA, WTDRD1, year, toddler, everything())
# create the survey design for HEI2020 day 1
HEI2020_NHANES_design_d1 = svydesign(
id = ~SDMVPSU,
strata = ~SDMVSTRA,
weight = ~WTDRD1,
data = HEI2020_NHANES_d1_2, #set up survey design on the full dataset #can restrict at time of analysis
nest = TRUE)